Pierre Gladieux 1, Darren Soanes 2, Sebastien Ravel 3, Marc-Henri Lebrun 4, Nick Talbot 2, Didier Tharreau 3, Elisabeth Fournier 1
1 INRA, UMR BGPI, Montpellier, France
2 University of Exeter, UK
3 CIRAD, UMR BGPI, Montpellier, France
4 INRA, UMR BIOGER, Thiverval-Grignon, France
Aims and objectives
Knowledge of pathogen populations, lineages, species, and their reproductive mode is an obligatory step to answer a host of questions common to all emerging diseases: is the disease due to a change in the environment that promotes disease, to the spread of a new pathogen, or to the emergence of a new lineage of an existing pathogen? What are the genomic changes and eco-evolutionary factors underlying disease emergence?
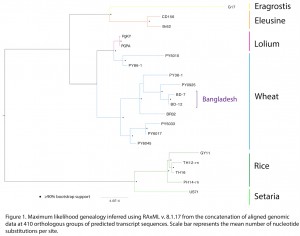
Here, our goal was to determine if « wheat blast » isolates recently collected in Bangladesh were related to Magnaporthe oryzae lineages infecting cereals and grasses using a phylogenomic approach. We also outline avenues for future research on the origins of wheat blast.
Materials
We used predicted consensus sequences obtained by aligning raw reads of Bangladeshi field samples No. 7 and 12 to the predicted transcriptome of Brazilian wheat-infecting Magnaporthe oryzae isolate BR32 (alignment: Kamoun lab @ TSL http://goo.gl/xpuD8L). We also used predicted transcript sequences extracted from the assembled genomic sequences of 20 Magnaporthe oryzae isolates collected on infected leaves of rice, wheat, Setaria millet, Eleusine spp., Lolium spp., and Eragrostis spp (Chiapello et al. 2015 GBE 7:2896-2912; Soanes & Talbot, Unpublished). This collection of isolates is representative of the known genetic diversity of M. oryzae. Except PY0925 and Bangladeshi isolates, for which no data are available, all M. oryzae isolates were pathogenic to their host plant of origin in controlled conditions.
Methods
We identified 4558 groups of sequences with orthologous relationships across the 22 transcriptomes (i.e. groups of sequences having exactly one gene for each isolate) using Proteinortho (Lechner et al. 2011 BMC Bioinformatics 12(1):124). We carried out alignments using MACSE (Ranwez et al. 2011 Plos One 6(9): e22594), with default parameters. We treated alignment gaps as missing data and kept only the 413 alignments with less than 10% sites with missing data, to reduce the impact of alignment errors. We excluded the regions corresponding to the first and last 16 codons (48bp) and treated ambiguities as missing data. We also excluded three transcript alignments for which Bangladeshi strains shared >5% sites with private doubletons, suggesting erroneous assignment of predicted sequences to BR32 transcripts.
Maximum likelihood phylogenetic inference was performed on the concatenated sequence of all orthologs (373,716 bp in total), using the GTRGAMMA model in RAxML v. 8.1.17 (Stamakis 2014 Bioinformatics 30: 1312) with 100 bootstrap replicates. The maximum likelihood genealogy was mid-point rooted along the longest branch, which was the branch connecting the Setaria millet- and rice-infecting lineages to other lineages, consistent with independent, unpublished, phylogenetic information (Gladieux et al.).
Results & Interpretation
The Bangladeshi isolates clustered, with high bootstrap support (>90%), with wheat infecting isolates of M. oryzae (Figure 1). The topology of the tree clearly shows that Bangladeshi isolates belongs to one of the two known M. oryzae wheat-infecting lineages. The findings presented here therefore indicate that the emergence of wheat blast in Bangladesh is likely caused by a wheat-infecting lineage of Magnaporthe oryzae, and is not a novel host lineage of the pathogen. The nesting of Bangladeshi samples within Brazilian wheat-infecting populations suggests a possible introduction in Bangladesh from a source in South America, which is the only continent where wheat-infecting populations of Magnaporthe oryzae have been reported.
Future research prospects
It remains difficult to identify the origin of fungal disease outbreaks and pathogen invasions because of limited baseline data concerning the prevalence of fungal diseases on wild hosts and the population structure of fungal pathogens infecting domesticated hosts (Taylor & Fisher 2003 Curr Op Microbiol 6:351-356; Taylor et al. 2006 Phil Trans B 361:1947-1963). Yet, identifying the biological (host species or variety) or geographical origin of pathogens is critical to understanding the initial steps leading to the emergence of a novel disease on a given host (Gladieux et al. 2015 Mol Ecol 24:1969-1986), and the fungal solution to the genetic paradox of invasions (i.e. the capacity for pathogens to adapt -for instance to new resistant varieties- despite limited genetic diversity).
The emergence of the wheat blast lineage of Magnaporthe oryzae in South America, and now in South Asia, is an unfortunate event with potentially disastrous consequences. However, it is also a unique opportunity to better understand the proximate (i.e. molecular) and ultimate (i.e. eco-evolutionary) determinants of pathogen colonization of new hosts and new areas. A first critical requirement for tackling this challenge was to determine the phylogenetic position of the causal agent of the novel disease. This was the purpose of the present study. Another critical requirement is to address the following questions regarding the origins of emerging populations:
What is the host species of origin of the wheat blast lineage of Magnaporthe oryzae? The difficulty is to distinguish infected wild hosts because of disease spillover from domesticated hosts, from the true wild relatives of pathogen populations that colonized agricultural habitats. Agrosystems provide a highly conducive environment for the maintenance of very large pathogen populations. In very large populations, virtually every possible mutation arises in every generation (Barton 2010 PLOS Genet 6(6): e1000987). One therefore expects the recurrent emergence of mutants that are capable to colonize wild relatives at the agricultural margins, even if they fail to establish self-sustainable populations (e.g. due to gene swamping; Lenormand 2002 Trends Ecol Evol 17: 183-189). It is important, therefore, to monitor the prevalence of disease and sample pathogen from wild relatives in areas located away from agrosystems. In addition, correct species identification of infected plants is critical, although particularly difficult for wild Poaceae. The emergence and spread of wheat blast reminds us how much it is important to characterize the diversity of cereal-associated Ascomycetes and their environmental reservoirs. Identifying the source of the wheat-infecting lineage of Magnaporthe oryzae will require the use of a speciation genomics analytical framework allowing the probabilistic comparison of colonization models, and the estimation of the time and demographics of lineage splitting events.
What is the geographical source of the Bangladeshi population of wheat-infecting Magnaporthe oryzae? Due to the strong genetic bottlenecks that occur during the invasion process, the genetic variability in the introduced pathogen population is expected to be a subset of that found in the source lineage from which it was derived. Thus, if source populations are extant in the geographical locations from which the pathogen was introduced, the geographical origin of invading populations can potentially be pinpointed to a particular host or region. Where population genetic structure is principally clonal, phylogenetic analysis can be useful to identify source populations, even if multiple independent introduction events occurred. In such asexual populations, the high degree of linkage disequilibrium among regions of the genome generates tree-like genome genealogies, and introduced isolates are therefore expected to cluster in highly supported clades nested within their source lineages. However, in recombining pathogens, identical genotypes are not expected, because each locus has a separate evolutionary history, and branches connecting individuals within recombining lineages are expected to be supported by low bootstrap values, precluding phylogenetic inference of the origins of disease outbreaks (Taylor et al. 1999 Ann Rev Phytopath 37:197-246; Fisher et al. 2002 PNAS 99:9067-9071). To identify the source of recombining pathogens, statistical methods such as Model-based Bayesian assignment methods or Approximate Bayesian computations are necessary. Approximate Bayesian computation, in particular, allows rigorous comparisons of complex scenarios modeling the genealogical relationships between populations and their geographic and demographic history. Previous population studies using microsatellite suggest that wheat blast populations may be recombining in Brazil (Maciel et al. 2014 Phytopath 104: 95-107), which could justify the use of methods accommodating recombining pathogens. Note that unlike the identification of the region and host of origin of emerging population, determining the taxonomic position of an isolate (as presented in this report) should be based on phylogenetic analyses, irrespective of the level of recombination (Taylor et al. 1999 Ann Rev Phytopath 37:197-246).
[May 8, 2016 update] Sequences of individuals genes, the RAXML dataset and RAXML outputs posted here: OWB-Report-data.