March 1, 2016. First news report of wheat blast in Bangladesh.
March 8, 2016. Tofazzal Islam and Sophien Kamoun discuss applying field pathogenomics to the wheat blast outbreak.
March 16, 2016. Tofazzal’s students collect samples from multiple locations in Bangladesh and store them in RNALater.
March 23, 2016. Field samples are delivered to The Sainsbury Lab in Norwich.
March 24-31, 2016. RNA extractions and library preparations in the Laboratory of Diane Saunders at TGAC and JIC.
April 8, 2016. Sequencing completed at TGAC. Thanks to Dan Swan and team for fast-tracking the samples.
April 14, 2016. Magnaporthe (Pyricularia) oryzae sequences confirmed based on analyses by Antoine Persoons and Joe Win. Similarity to Br32 wheat blast strain noted.
April 18, 2016. Open Wheat Blast goes live! All sequence data is freely available to use without any restrictions. Nick Talbot’s group at Exeter simultaneously releases the genome sequences of 14 Brazilian wheat blast isolates.
April 21, 2016. Hon. Matia Chowdhury, Minister of Agriculture, People’s Republic of Bangladesh receives Tofazzal Islam and team, and recognizes Open Wheat Blast initiative.
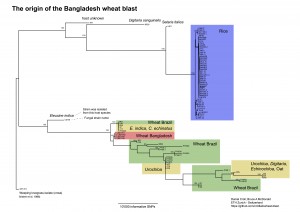
April 27, 2016. Daniel Croll and Bruce McDonald, ETH Zurich, post in Github an analysis on the origin of wheat blast in Bangladesh. They conclude that the pathogen was most likely introduced from South America.
April 27, 2016. Ewen Callaway‘s article in Nature News “Scientists race to determine origin of Bangladesh outbreak, which they warn could spread farther afield.”
Time from field collection to genome analysis: 6 weeks.
Read more about field pathogenomics here.